Self-supervised denoising for multimodal structured illumination microscopy enables long-term super-resolution live-cell imaging
doi: 10.1186/s43074-024-00121-y
Self-supervised denoising for multimodal structured illumination microscopy enables long-term super-resolution live-cell imaging
-
Abstract:
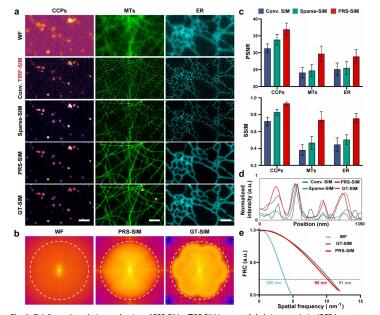
Detection noise significantly degrades the quality of structured illumination microscopy (SIM) images, especially under low-light conditions. Although supervised learning based denoising methods have shown prominent advances in eliminating the noise-induced artifacts, the requirement of a large amount of high-quality training data severely limits their applications. Here we developed a pixel-realignment-based self-supervised denoising framework for SIM (PRS-SIM) that trains an SIM image denoiser with only noisy data and substantially removes the reconstruction artifacts. We demonstrated that PRS-SIM generates artifact-free images with 20-fold less fluorescence than ordinary imaging conditions while achieving comparable super-resolution capability to the ground truth (GT). Moreover, we developed an easy-to-use plugin that enables both training and implementation of PRS-SIM for multimodal SIM platforms including 2D/3D and linear/nonlinear SIM. With PRS-SIM, we achieved long-term super-resolution live-cell imaging of various vulnerable bioprocesses, revealing the clustered distribution of Clathrin-coated pits and detailed interaction dynamics of multiple organelles and the cytoskeleton.
-
Key words:
- Engineering" /
- " data-track="click" data-track-action="view keyword" data-track-label="link">Super-resolution microscopy /
- Engineering" /
- " data-track="click" data-track-action="view keyword" data-track-label="link">Structured illumination microscopy /
- Engineering" /
- " data-track="click" data-track-action="view keyword" data-track-label="link">Deep learning /
- Engineering" /
- " data-track="click" data-track-action="view keyword" data-track-label="link">Self-supervised learning /
- Engineering" /
- " data-track="click" data-track-action="view keyword" data-track-label="link">Live-cell imaging
-
[1] Gustafsson MG. Surpassing the lateral resolution limit by a factor of two using structured illumination microscopy. J Microsc. 2000;198:82–7. [2] Gustafsson MG, Shao L, Carlton PM, et al. Three-dimensional resolution doubling in wide-field fluorescence microscopy by structured illumination. Biophys J. 2008;94:4957–70. [3] Karras C, Smedh M, Förster R, et al. Successful optimization of reconstruction parameters in structured illumination microscopy–a practical guide. Opt Commun. 2019;436:69–75. [4] Demmerle J, Innocent C, North AJ, et al. Strategic and practical guidelines for successful structured illumination microscopy. Nat Protoc. 2017;12:988–1010. [5] Kner P, Chhun BB, Griffis ER, et al. Super-resolution video microscopy of live cells by structured illumination. Nat Methods. 2009;6:339–42. [6] Li D, Shao L, Chen B-C, et al. Extended-resolution structured illumination imaging of endocytic and cytoskeletal dynamics. Science. 2015;349:aab3500. [7] Guo Y, Li D, Zhang S, et al. Visualizing intracellular organelle and cytoskeletal interactions at nanoscale resolution on millisecond timescales. Cell. 2018;175:1430-1442. e1417. [8] Chu K, McMillan PJ, Smith ZJ, et al. Image reconstruction for structured-illumination microscopy with low signal level. Opt Express. 2014;22:8687–702. [9] Labouesse S, Negash A, Idier J, et al. Joint reconstruction strategy for structured illumination microscopy with unknown illuminations. IEEE Trans Image Process. 2017;26:2480–93. [10] Smith CS, Slotman JA, Schermelleh L, et al. Structured illumination microscopy with noise-controlled image reconstructions. Nat Methods. 2021;18:821–8. [11] Wen G, Li S, Wang L, et al. High-fidelity structured illumination microscopy by point-spread-function engineering. Light: Sci Appl. 2021;10:70. [12] Hagen N, Gao L, Tkaczyk TS. Quantitative sectioning and noise analysis for structured illumination microscopy. Opt Express. 2012;20:403–13. [13] Wu Y, Shroff H. Faster, sharper, and deeper: structured illumination microscopy for biological imaging. Nat Methods. 2018;15:1011–9. [14] Liu SB, Xie BK, Yuan RY, et al. Deep learning enables parallel camera with enhanced-resolution and computational zoom imaging. PhotoniX. 2023;4:1–20. [15] Wu Y, Han X, Su Y, et al. Multiview confocal super-resolution microscopy. Nature. 2021;600:279–84. [16] Kreiss L, Jiang S, Li X, et al. Digital staining in optical microscopy using deep learning - a review. PhotoniX. 2023;4:1–32. [17] Qiao C, Li D, Guo Y, et al. Evaluation and development of deep neural networks for image super-resolution in optical microscopy. Nat Methods. 2021;18:194–202. [18] Huang B, Li J, Yao B, et al. Enhancing image resolution of confocal fluorescence microscopy with deep learning. PhotoniX. 2023;4:1–22. [19] Wang H, Rivenson Y, Jin Y, et al. Deep learning enables cross-modality super-resolution in fluorescence microscopy. Nat Methods. 2019;16:103–10. [20] Jin L, Liu B, Zhao F, et al. Deep learning enables structured illumination microscopy with low light levels and enhanced speed. Nat Commun. 2020;11:1–7. [21] Qiao C, Chen X, Zhang S, et al. 3D structured illumination microscopy via channel attention generative adversarial network. IEEE J Sel Top Quantum Electron. 2021;27:1–11. [22] Shah ZH, Müller M, Wang TC, et al. Deep-learning based denoising and reconstruction of super-resolution structured illumination microscopy images. Photonics Res. 2021;9:B168-181. [23] Yang W, Zhang X, Tian Y, et al. Deep learning for single image super-resolution: a brief review. IEEE Trans Multimedia. 2019;21:3106–21. [24] Dong C, Loy CC, He K, Tang X. Image super-resolution using deep convolutional networks. IEEE Trans Pattern Anal Mach Intell. 2015;38:295–307. [25] Qiao C, Li D, Liu Y, et al. Rationalized deep learning super-resolution microscopy for sustained live imaging of rapid subcellular processes. Nat Biotechnol. 2023;41:1–11. [26] Rahaman N, Baratin A, Arpit D, Draxler F, Lin M, Hamprecht F, Bengio Y, Courville A. On the spectral bias of neural networks. International Conference on Machine Learning. 2019. p. 5301–5310. PMLR. [27] Xu ZQ, Zhang Y, Luo T, Xiao Y, Ma Z. Frequency principle: Fourier analysis sheds light on deep neural networks. arXiv preprint arXiv:1901.06523. 2019. [28] Zhang K, Zuo W, Chen Y, et al. Beyond a gaussian denoiser: residual learning of deep cnn for image denoising. IEEE Trans Image Process. 2017;26:3142–55. [29] Zhang K, Zuo W, Zhang L. FFDNet: toward a fast and flexible solution for CNN-based image denoising. IEEE Trans Image Process. 2018;27:4608–22. [30] Weigert M, Schmidt U, Boothe T, et al. Content-aware image restoration: pushing the limits of fluorescence microscopy. Nat Methods. 2018;15:1090–7. [31] Chen J, Sasaki H, Lai H, et al. Three-dimensional residual channel attention networks denoise and sharpen fluorescence microscopy image volumes. Nat Methods. 2021;18:678–87. [32] Chen B-C, Legant WR, Wang K, et al. Lattice light-sheet microscopy: imaging molecules to embryos at high spatiotemporal resolution. Science. 2014;346: 1257998. [33] Rego EH, Shao L, Macklin JJ, et al. Nonlinear structured-illumination microscopy with a photoswitchable protein reveals cellular structures at 50-nm resolution. Proceedings of the National Academy of Sciences. 2012;109:E135–43. [34] Gustafsson MG. Nonlinear structured-illumination microscopy: wide-field fluorescence imaging with theoretically unlimited resolution. Proceedings of the National Academy of Sciences. 2005;102:13081–6. [35] Huang T, Li S, Jia X, et al. Neighbor2neighbor: a self-supervised framework for deep image denoising. IEEE Trans Image Process. 2022;31:4023–38. [36] Lequyer J, Philip R, Sharma A, et al. A fast blind zero-shot denoiser. Nat Mach Intell. 2022;4:1–11. [37] Qiao C, Li D. BioSR: a biological image dataset for super-resolution microscopy. Figshare; 2022. https://figshare.com/articles/dataset/BioSR/13264793. [38] Nieuwenhuizen RP, Lidke KA, Bates M, et al. Measuring image resolution in optical nanoscopy. Nat Methods. 2013;10:557–62. [39] Krull A, Buchholz TO, Jug F. Noise2void-learning denoising from single noisy images. Proceedings of the IEEE/CVF conference on computer vision and pattern recognition. 2019. p. 2129–2137. [40] Prakash M, Delbracio M, Milanfar P, Jug F. Interpretable unsupervised diversity denoising and artefact removal. arXiv preprint arXiv:2104.01374. 2021. [41] Pang T, Zheng H, Quan Y, Ji H. Recorrupted-to-recorrupted: Unsupervised deep learning for image denoising. Proceedings of the IEEE/CVF conference on computer vision and pattern recognition 2021. p. 2043–2052. [42] Wang Z, Liu J, Li G, Han H. Blind2unblind: Self-supervised image denoising with visible blind spots. Proceedings of the IEEE/CVF conference on computer vision and pattern recognition. 2022. p. 2027–2036. [43] Zhang G, Li X, Zhang Y, et al. Bio-friendly long-term subcellular dynamic recording by self-supervised image enhancement microscopy. Nat Methods. 2023;20:1957–70. [44] Zhao W, Zhao S, Li L, et al. Sparse deconvolution improves the resolution of live-cell super-resolution fluorescence microscopy. Nat Biotechnol. 2022;40:606–17. [45] Wang Z, Zhao T, Hao H, et al. High-speed image reconstruction for optically sectioned, super-resolution structured illumination microscopy. Adv Photonics. 2022;4:026003–026003. [46] Wen G, Li S, Liang Y, et al. Spectrum-optimized direct image reconstruction of super-resolution structured illumination microscopy. PhotoniX. 2023;4:1–18. [47] Lehtinen J, Munkberg J, Hasselgren J, Laine S, Karras T, Aittala M, Aila T. Noise2Noise: Learning image restoration without clean data. arXiv preprint arXiv:1803.04189. 2018. [48] Ronneberger O, Fischer P, Brox T. U-Net: Convolutional networks for biomedical image segmentation. arXiv preprint arXiv:1505.04597. 2015. [49] Ehrlich M, Boll W, Van Oijen A, et al. Endocytosis by random initiation and stabilization of clathrin-coated pits. Cell. 2004;118:591–605. [50] Godlee C, Kaksonen M. From uncertain beginnings: initiation mechanisms of clathrin-mediated endocytosis. J Cell Biol. 2013;203:717–25. [51] Cureton DK, Massol RH, Saffarian S, et al. Vesicular stomatitis virus enters cells through vesicles incompletely coated with clathrin that depend upon actin for internalization. PLoS Pathog. 2009;5: e1000394. [52] Arganda-Carreras I, Kaynig V, Rueden C, et al. Trainable Weka Segmentation: a machine learning tool for microscopy pixel classification. Bioinformatics. 2017;33:2424–6. [53] Li X, Zhang G, Wu J, et al. Reinforcing neuron extraction and spike inference in calcium imaging using deep self-supervised denoising. Nat Methods. 2021;18:1395–400. [54] Li X, Li Y, Zhou Y, et al. Real-time denoising enables high-sensitivity fluorescence time-lapse imaging beyond the shot-noise limit. Nat Biotechnol. 2023;41:282–92. [55] Schindelin J, Arganda-Carreras I, Frise E, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9:676–82. [56] Qian J, Cao Y, Bi Y, et al. Structured illumination microscopy based on principal component analysis. eLight. 2023;3:4. [57] Wang Z, Zhao T, Cai Y, et al. Rapid, artifact-reduced, image reconstruction for super-resolution structured illumination microscopy. The Innovation. 2023;4:100425. [58] Hampson KM, Turcotte R, Miller DT, et al. Adaptive optics for high-resolution imaging. Nat Reviews Methods Primers. 2021;1:1–26. [59] Zhou Z, Huang J, Li X, et al. Adaptive optical microscopy via virtual-imaging-assisted wavefront sensing for high-resolution tissue imaging. PhotoniX. 2022;3:1–20. [60] Turcotte R, Liang Y, Tanimoto M, et al. Dynamic super-resolution structured illumination imaging in the living brain. Proc Natl Acad Sci. 2019;116:9586–91. [61] Zhang Y, Wang Y, Wang M, et al. Multi-focus light-field microscopy for high-speed large-volume imaging. PhotoniX. 2022;3:1–20. [62] Müller M, Mönkemöller V, Hennig S, et al. Open-source image reconstruction of super-resolution structured illumination microscopy data in ImageJ. Nat Commun. 2016;7:1–6. [63] Cao R, Li Y, Chen X, et al. Open-3DSIM: an open-source three-dimensional structured illumination microscopy reconstruction platform. Nat Methods. 2023;20:1183–6. [64] Zhang Y, Li K, Li K, Wang L, Zhong B, Fu Y. Image super-resolution using very deep residual channel attention networks. Proceedings of the European conference on computer vision (ECCV). 2018. p. 286–301. [65] Huang G, Liu Z, Pleiss G, et al. Convolutional networks with dense connectivity. IEEE Trans Pattern Anal Mach Intell. 2019;44:8704–16. [66] Ehlschlaeger CR, Shortridge AM, Goodchild MF. Visualizing spatial data uncertainty using animation. Computers Geosci. 1997;23:387–95. [67] Tinevez J-Y, Perry N, Schindelin J, et al. TrackMate: an open and extensible platform for single-particle tracking. Methods. 2017;115:80–90. [68] Manders E, Verbeek F, Aten J. Measurement of co-localization of objects in dual‐colour confocal images. J Microsc. 1993;169:375–82. [69] Sanjana NE, Cong L, Zhou Y, et al. A transcription activator-like effector toolbox for genome engineering. Nat Protoc. 2012;7:171–92.









 下载:
下载: